
Onconomics Intranet
| Home | |
| Press Releases | |
| Human Resources Department | |
| Sequencing Department | |
| Bioinformatics Department | |
| External Links | |
| Glossary | |
Cycle Sequencing Protocol
Step 1: A DNA sequence of interest is first amplified using the polymerase chain reaction (PCR).
- Add to reaction tube:
- Double-stranded DNA sequence of interest
- Taq DNA polymerase (a heat-resistant enzyme that catalyzes the addition of deoxynucleotides)
- A primer (a short single-stranded DNA sequence that base pairs to the DNA sequence of interest and serves as a site from which to add complementary bases.)
- A supply of the four deoxynucleotides
- Heat to separate DNA strands
- Cool to allow primers to bind
- Taq DNA polymerase extends the 3' end of each primer to produce double-stranded DNA molecules
- Repeat for 20 cycles.
Step 2: The amplified DNA strands are subjected to the DNA sequencing reactions.
- Add to reaction tubes:
- Amplified DNA strand from Step 1
- Taq DNA polymerase
- Primer
- All four deoxynucleotides
- Dideoxynucleotides (human-made nucleotides that can not form covalent bonds to another nucleotide and are labeled with a fluorescent dye. The addition of a dideoxynucleotide terminates the growing DNA strand)
- Run through 20 30 cycles of heating and cooling.
Step 3: The results of the sequencing reactions are separated by capillary gel electrophoresis and visualized using fluorescent dyes.
- Load the contents of the sequencing reactions into capillary tubes. We use capillary tubes made of fused silica (glass) and coated on the outside with polyimide (for strength). The tubes have an internal diameter of 100 micrometers and are 100 cm in length. The tubes contain a gel solution which serves as a sieving matrix. Each of our electrophoresis machines holds 96 capillary tubes.
- Begin electrophoresis.
- The DNA strands migrate, and therefore separate from each other, according to size. The smallest strand travels the fastest and is detected first.
- A laser beam directed at the bottom of the capillary tubes excites the fluorescent-labeled dideoxynucleotide which is positioned at the end of the DNA strand.
- A photodetector measures the light emitted by each dideoxynucleotide as it passes through the laser beam.
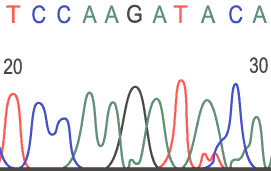
- The
computer records each wavelength of light and generates an electropherogram
with colored peaks representing each wavelength. The terminal base
(the dideoxynucleotides) of the shortest fragment is the first base
in the electropherogram. The colors of each base are listed below:
- Guanine (G) yellow (the tracings on the chart paper use black instead of yellow)
- Cytosine (C) blue
- Adenine (A) green
- Thymine (T) red
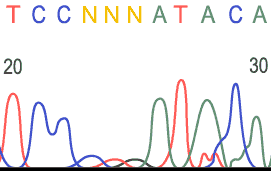
- Unknown base = N
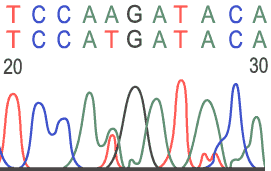
Sample Electropherograms
Normal Sequence:
Heterozygous Base:
Unreadable Region:
Home
Copyright © 2003, Onconomics Corporation
![]()